Projects
Label combination in cause-of-death analysis (2023)
In 2023, I work in Prof. Zehang Richard Li at UCSC. We focus on cause-of-death analysis where post-training label combination is important if we want to yield appropriately ambiguous results without sacrificing essential information. Existing label combination methods are designed exclusively for classification tasks, so we need to extend them to tasks with a different loss like cause-of-death analysis. Our key design is to formulate the label combination problem as a new optimization problem with a proposed regularizer to control the combination degree.
Microbiota and antibiotic resistance genes at human-pig-soil interface (2022-2023)
In September 2021, I joined Prof. Xiaokui Guo and Chang Liu’s lab to lead this project. 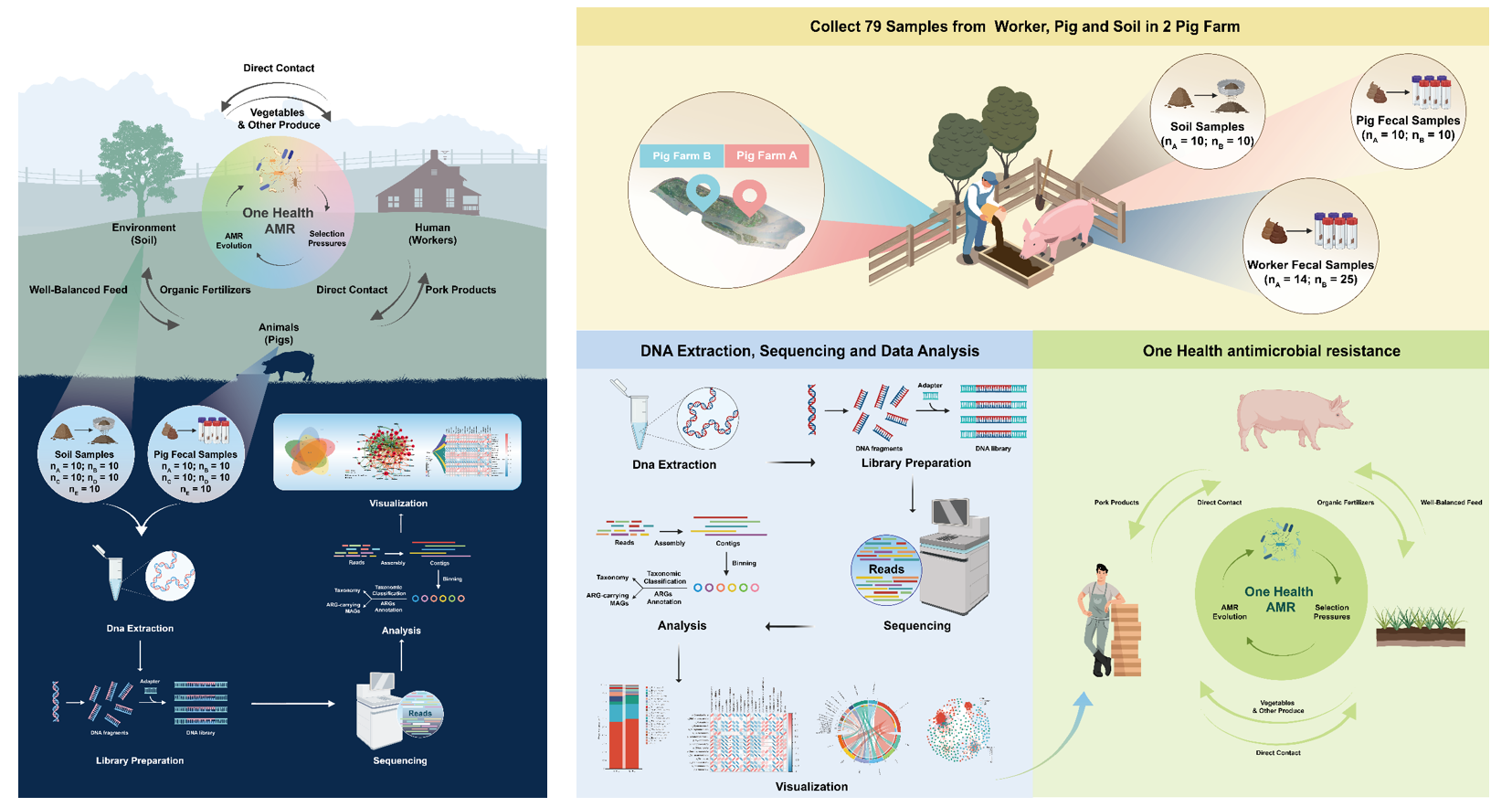 Mapping microbial connectivity of people, animals and their environment. [LeftFig-HighRes] [RightFig-HighRes]
Mapping microbial connectivity of people, animals and their environment. [LeftFig-HighRes] [RightFig-HighRes]
Swine farming is a widespread agricultural practice that produces a substantial amount of antibiotics and genes associated with antibiotic resistance in swine manure. The presence of these antibiotic resistance genes (ARGs) poses a potential threat to the environment and human health. In this project we propose a unique study design and epidemiological environment to sample workers, pigs and soil from 5 farms and use Index 2 × 150 paired-end sequencing through Illumina NovaSeq 6000 for raw data. Our analytical pipeline includes genome assembly programs, taxonomic classifiers, read mappers, and pathway reconstruction tools. We also implement an improved binning strategy, retrieving both short-read and long-read contigs belonging to the same genome. This hybrid assembly approach has lower error rates, produces more contiguous assemblies, generates metagenomic assembled genomes, and allows for the reconstruction of near-complete genomes.
A survey of antibiotic resistance from a One Health perspective (2021-2022)
 Transmission of antibiotic resistance genes at the human-animal-environment interfaces. [LeftFig-HighRes] [RightFig-HighRes]
Transmission of antibiotic resistance genes at the human-animal-environment interfaces. [LeftFig-HighRes] [RightFig-HighRes]
Relevant publications:
- Jing Qian, Zheyuan Wu, Yongzhang Zhu, Chang Liu. One Health: a holistic approach for food safety in livestock. Science in One Health. 2023:100015. [View]
- Jing Qian, Zheyuan Wu, Xiaokui Guo, Chang Liu. Antibiotic-resistant microbes,antibiotic resistance genes and One Health. Microbiology China. 2022, 49(10): 4412-4424. [View]
- Zheyuan Wu, Jing Qian, Qingtian Li, Chang Liu, Xiaokui Guo. Progress in the Study of Antibiotic Resistance Genes Based on Metagenomic Next Generation Sequencing. Chinese Journal of Microecology. 2023,35(06):726-729+735 [View]
In the entire ecosystem, rich diversities of microorganisms exist at the human-animal interface, some of which cause human and/or animal diseases. There are 58% of human pathogens that are zoonotic and cause diseases in both humans and animals. The food safety of livestock is a critical issue between animals and humans due to their complex interactions. Pathogens have the potential to spread at every stage of the animal food handling process, including breeding, processing, packaging, storage, transportation, marketing and consumption. In addition, application of the antibiotic usage in domestic animals is a controversial issue because, while they can combat food-borne zoonotic pathogens and promote animal growth and productivity, they can also lead to the transmission of antibiotic-resistant microorganisms and antibiotic-resistant genes across species and habitats. Coevolution of microbiomes may occur in humans and animals as well which may alter the structure of the human microbiome through animal food consumption. One Health is a holistic approach to systematically understand the complex relationships among humans, animals and environments which may provide effective countermeasures to solve food safety problems aforementioned. This paper depicts the main pathogen spectrum of livestock and animal products, summarizes the flow of antibiotic-resistant bacteria and genes between humans and livestock along the food-chain production, and the correlation of their microbiome is reviewed as well to advocate for deeper interdisciplinary communication and collaboration among researchers in medicine, epidemiology, veterinary medicine and ecology to promote One Health approaches to address the global food safety challenges.
